Information Governance in OpenPathology
This page provides of a summary of our Data Protection Impact Assessment (DPIA), which has been signed off with our division's Information Compliance team.
How will we obtain data?
Data will be collected from NHS pathology laboratories based in NHS Hospital Trusts.
This data is stored by each Trust in their own LIMS (pathology database), which are typically in different formats. When a pathology request comes in from primary or secondary care, the request is stored in the LIMS. Every LIMS is different. In each case, we will ask for a monthly extract which covers the requestor, their location, the test(s) requested, an internal identifier for the sample, an internal (hospital) identifier for the patient, sex, and age.
We will contact each Trust separately and follow their Data Sharing procedures to agree an extract of this database. When an agreement is signed, a LIMS operator will carry out a database extract and provide the data to us in CSV or Excel format.
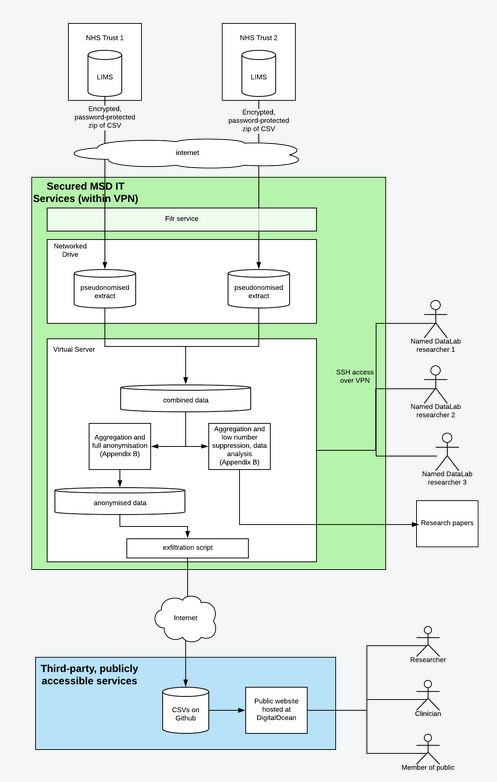
Data will be transferred from Trusts via the Filr service provided by the University’s Medical Sciences Division (MSD) IT service, and then transferred to the virtual server over SSH. These services are provided by MSD IT, and meet the University’s baseline standard.
How will we store and process the data?
The data will be stored and initially processed on a network drive accessed via a secure virtual hosted server, accessible to 3 named individuals in our team. The server will be provided and managed by the University’s Medical Sciences Division (MSD) IT service.
The individuals permitted access will process the data to further anonymise and aggregate it into two formats, within the secure area (marked Secured MSD IT Services in Appendix A).
- The first format will be entirely anonymised, suitable for publication in full as data.
- The second format will be pseudonymised (prior to us receiving the data) and aggregated with low-number suppression for incorporation into research publications; this will contain high-level identifiable data in the form of sex and age band, grouped by these and aggregated by test results, and with low numbers (N < 5) suppressed.
Prior to publication, anonymisation of both formats will be categorised by the SIRO as per the Departmental Anonymisation policy. Examples are given in Appendix B.
The fully anonymised data will leave the secure area over SSH and will be processed on third-party servers to perform further statistical analyses and visualisations to help audit and inform pathology activities by requesting clinicians. This data will be freely available to the public, with the agreement of each Trust, and as such no longer subject to strict security controls.
Anonymised extracts of the aggregated, pseudonymised data will be published in data tables in individual research papers.
Purpose of data processing
- To provide GPs simple, actionable feedback on their request rates and results data, allowing them to e.g. identify outliers for targeted improvement, and investigate those responding quickly or slowly to changes in cost and evidence. This will improve patient outcomes by helping address under- and over-testing rates
- By creating a tool which could be rolled out nationally we save local NHS teams time, effort and money in building these tools themselves.
- To document variation in care in relation to pathology testing, more widely than our recent similar studies.
Our lawful basis is GDPR articles 6.1.e public interest and 9.2.j research. See Stage 2 part 4, above (cost saving for NHS, improving patient outcomes)
Transparency
We will add a page on our website for individuals whose data may be included, providing the following transparency information:
- The purposes for which their data will be processed?
- The people or organisations their data will be shared with?
- The lawful basis for processing their data?
- Any international transfers of their personal data
- When their personal data will be erased?
- Their rights under GDPR?
Data retention
Data will be kept for 5 years, in order to provide time trends analyses.
Ethics
There is no need for ethics approval for the project at present, as this was classed by the Ethics Team as a “service development” project. We will seek appropriate ethical approvals for research projects as required.
Data flow diagram
Anonymisation schedule
|
Field name |
Purpose |
Example aggregation for research publication |
Included in anonymised dataset? |
|---|---|---|---|
|
Location |
Primary aggregation and analysis level, e.g. number of tests per GP practice |
Not aggregated: data grouped by this field |
Yes: data grouped by this field |
|
Test identifier |
Compare tests between locations |
Not aggregated: data grouped by this field |
Yes: data grouped by this field |
|
Requestor |
Compare number of requests per clinician |
Count of requestor per location or test |
No |
|
Test results |
Compare results that have positive or negative outcomes |
Count of positive or negative results; median of results; deciles of results |
Yes: count of positive and negative results, summary statistics of result |
|
Internal patient identifier |
Compare number of repeat or new tests over time per-patient |
Count of repeat or novel tests per-patient per location and per test |
No |
|
Patient sex |
Filter tests by sex (some tests have different meanings for different sexes); compare tests by sex |
Count of positive test results per age band, per sex |
No |
|
Patient age |
Filter tests by age band; compare results by age band |
Count of positive test results per age band, per location |
No |
Where a Lab provides an internal patient identifier, this will be either a randomised key or a hospital unit number. We will never accept data including an NHS number. Hospital patient identifiers are always internal to the Trust and the ability to match these to patients will not be held by this project.
Example extract of anonymised dataset for full publication:
Note we would like to avoid low number suppression in this dataset as it will make working with the data much easier and allow better visualisations of the data. In terms of a deidentification attack, we have not been able to think of a way a person could be identified from a test and GP practice, even in combination with other sources. An open data NHS dataset we work with routinely contains data at a similar level (GP practice and drug) and does not have low number suppression, so we are familiar with thinking about these risks.
|
Location |
Test |
Within reference range |
Outside reference range |
Median value |
|---|---|---|---|---|
|
P90112 |
WBC |
42 |
5 |
302.5 |
|
P90113 |
WBC |
32 |
3 |
250.6 |
|
P90114 |
WBC |
22 |
52 |
410.2 |
|
P90112 |
HBA |
3 |
1 |
3005.9 |
|
P90113 |
HBA |
10 |
5 |
3011.0 |
|
P90114 |
HBA |
20 |
12 |
4100.5 |
Example extract of aggregated data for research publication, with low number suppression:
As this data includes age bands, to be cautious, we will suppress low numbers to reduce the risk of reidentification.
|
Location |
Test |
Age band |
% first test |
Within reference range |
Outside reference range |
|---|---|---|---|---|---|
|
99P |
WBC |
0-5 |
100 |
[suppressed] |
[suppressed] |
|
99P |
WBC |
6-10 |
99.5 |
34 |
11 |
|
99P |
WBC |
11-20 |
90.1 |
50 |
12 |
|
99P |
WBC |
21-30 |
85.0 |
3013 |
105 |
|
99P |
WBC |
31-40 |
67.5 |
3406 |
210 |
|
99P |
WBC |
41-50 |
48.8 |
3678 |
581 |